2.1 Filtering the results An additional option is the filtering of computationally predicted target results on lncRNAs. The user can filter the results using an arbitrary microT threshold, fine-tuning prediction sensitivity and precision levels. The default threshold value is 0.8 which provides adequate sensitivity and precision for most analyses. The results can be additionally refined by an lncRNA tissue specificity filter. 2.2 Interactions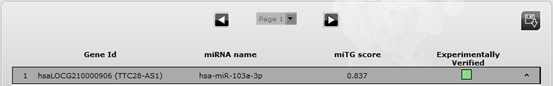 Each entry presents general information, regarding the lncRNAs and miRNAs participating in each interaction, along with the predicted score, and an indication of whether each interaction has also been experimentally verified. A green (predicted & validated) or grey (predicted) colored indicator is utilized to this end. Upon clicking the indicator, direct access to the corresponding section in the experimental module is provided. By expanding each entry, the user can access several informative sections. Note that the queried results can be downloaded by clicking the appropriate button in the result space. 2.3 Genes The Gene details section supports links and general information regarding the lncRNAs, such as tissue expression. 2.4 microRNAs The section miRNA details provides a hyperlink to the relevant miRBase entry, shows the corresponding miRBase id, miRNA sequence and also presents a tag cloud with Medical Subject Headings (MeSH) terms derived from MedLine publications. 2.5 MREs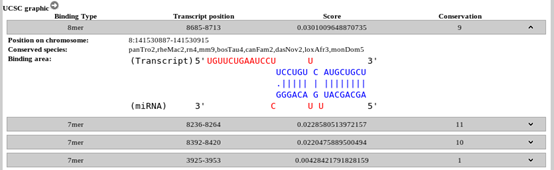 Each miRNA-lncRNA interaction has also one expandable entry with general information per predicted MRE, such as the binding type, MRE location on the transcript, the prediction score and the number of species that the MRE was found to be conserved. Upon expanding the MRE entry, the user can access additional information regarding the position of the MRE on the chromosome, the species that the MRE is conserved and a graphical representation of the binding area. An additional feature of the Predicted Module is the ability to visualize the interactions in the UCSC genome browser. Upon clicking the relevant button, a graphical representation appears containing all the MREs, of the selected lncRNA, whose interaction score is above 0.8. This threshold has been deliberately chosen in order to produce a graphic that is not crowded and can be efficiently interpreted by the user. |