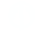| | | DIANA-web server Taverna Plug-in. | | |
The use of the plugin is highly suggested, since it provides optimized use of the DIANA-web server and databases.
Install DIANA-LAB plugin in Taverna:
- Download and install Taverna 2.3 or Taverna 2.5
- Start Taverna
- Dowload one of the files below depending on the Taverna version you are using:
Taverna 2.3 or Taverna 2.5
Then, unzip the file and follow the instructions on the Readme file.
- Restart Taverna
The DIANA-LAB: http://62.217.127.8/DianaTools services plugin should now be displayed with others local tools under the section "Available Services".
Description of the Taverna Plugin available Services
DIANA-microT-ANN (v4) service
By Using DIANA-Taverna Plugin, the user can directly access the web server and identify microRNAs (miRNAs) predicted to target selected genes OR find gene targets of selected miRNAs. The input/output ports of the DIANA-microT-ANN (v4) service are described below:
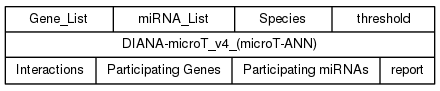
The user has to specify the input ports of the DIANA-microT_v4 (microT-ANN) service in the Taverna plugin:
- Gene_List: DIANA-microT_v4 can be queried using a gene name/identifier, or with a list of gene names/identifiers (gene names OR Ensembl 69 gene ids separated by a carriage return / newline character). Example value: FBgn0086758.
- miRNA_List: DIANA-microT_v4 can be queried with a miRNA name/identifier, or with a list of miRNA names/identifiers (miRNA names OR MIMAT ids are separated by a carriage return / newline character). Example value: dme-let-7-5p.
- Species: Species supported by the web server in org code, alias or full name format, eg hsa, human or homo sapiens
- threshold: A prediction score cut off value for presented predictions, ranging from 0.3 to 1. If no threshold is defined by the user, prediction results are provided for a default value of 0.7.
The output ports (provided results) of the DIANA-microT_v4 (microT-ANN) service in the Taverna plugin are presented below:
- Interactions: Predicted microRNA-gene interactions
- Participating Genes: Ensembl v69 gene ids of the targets present in the predicted interactions.
- Participating miRNAs: mature miRNA names (miRBase v18) of the miRNAs taking part in the predicted interactions.
- report: General information about the provided results
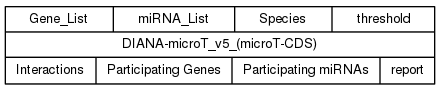
DIANA-microT-CDS (v5) Service
DIANA-microT-CDS service follows the exact same syntax as DIANA-microT v4, presenting the same input/output ports as shown below.
DIANA-TarBase v2.0 Service
This is a service to query directly DIANA-TarBase v6.0 the largest available database indexing manually curated experimentally validated miRNA-gene interactions. The user can query using a gene name / ENSEMBL gene ID (preferred) OR miRNA name (miRBase 18+ nomenclature) / MIMAT ID.
The input/output ports of the DIANA-TarBase v6.0 service are described below:

The user has to specify at least one of the input ports of the DIANA-TarBase v6.0 service in the Taverna plugin:
- Gene_List: DIANA-TarBase v6.0 can be queried with a gene name/identifier, or with a list of gene names/identifiers (gene names OR Ensembl 69 gene ids separated by a carriage return / newline character). Example value: TUSC2.
- miRNA_List: DIANA-TarBase v6.0 can be queried with a miRNA name/identifier, or with a list of miRNA names/identifiers (miRNA names OR MIMAT ids are separated by a carriage return / newline character). Example value: hsa-let-7a-5p.
- Species: Species supported by the web server in org code, alias or full name format, eg hsa, human or homo sapiens
DIANA-miRPath v2.1 Web Service
This service queries DIANA-miRPath server and identifies significantly targeted pathways by the selected miRNA(s). The miRNA-gene interactions can be derived directly from TarBase, or can be computationally predicted using DIANA-microT-CDS. In case where more than one miRNAs are queried, DIANA miRPath identifies significantly targeted pathways by assessing the combinatorial effect of the selected miRNAs. The input/output ports of the DIANA-miRPath service are described below:

- Gene filtering: A url to a file containing a list of gene name/identifiers separated by newline character. The user can upload to a public url a predefined list of genes that are expressed in investigated tissues. DIANA will filter all miRNA targets based on this list and will use only the expressed subset of genes for the pathway enrichment analysis.
- List miRNA - Validation: DIANA-miRPath can be queried with a miRNA name/identifier followed by validation method Tarbase or microT-CDS, or with a list of miRNA names/identifiers each one accompanied by a validation method (miRNA names OR MIMAT ids –validation methods pairs are separated by a carriage return / newline character). miRNA name – validation method terms can be separated by comma, space or tab characters. If no validation method is provided for a miRNA then the service enables the microT-CDS as a default value. Example value: hsa-mir-125b-5p Tarbase.
- Merging Genes: union/intersection
- Merging Pathways: union/intersection
- Species: Example values: human, mouse
- Statistics Conservative: true/false
- Statistics FDR: true/false
- threshold_microT: A cut off value for presented predictions (when microT-CDS is utilized as a validation method), ranging from 0.3 to 1.
The output ports (provided results) of the DIANA-microT_v4 (microT-ANN) service in the Taverna plugin are presented below:
- miRNAs-Pathways: DIANA-miRPath results, containing information such as Pathway KEGGid, Pathway description, Number of Associated genes, Gene Names, pValue Participating miRNAs.
- Pathways: A list with the pathways KeggIds significantly targeted by the selected miRNA(s).
- report: General information about the provided results
Example Workflows
A series of ready-made workflows have been prepared, which can be used as standalone analysis workflow, as a foundation for custom pipelines OR can be inserted into pre-existing pipelines.
These pipelines can be utilized to analyze user data derived from small scale and high throughput experiments directly from the DIANA-microT web server interface, without the necessity to install or implement any kind of software. In all workflows the user has to specify the species and two lists of differentially expressed mRNAs (microarray/RNA-Seq) and miRNAs (microarray/sRNA-Seq) respectively. The gene list has to contain Ensembl gene identifiers, while the miRNA list should be composed of miRNA names/identifiers according to miRBase nomenclature. miRNA and gene identifiers can optionally be followed by fold change values. In this case, the workflows automatically matches suppressed genes are with overexpressed miRNAs (and vice versa).
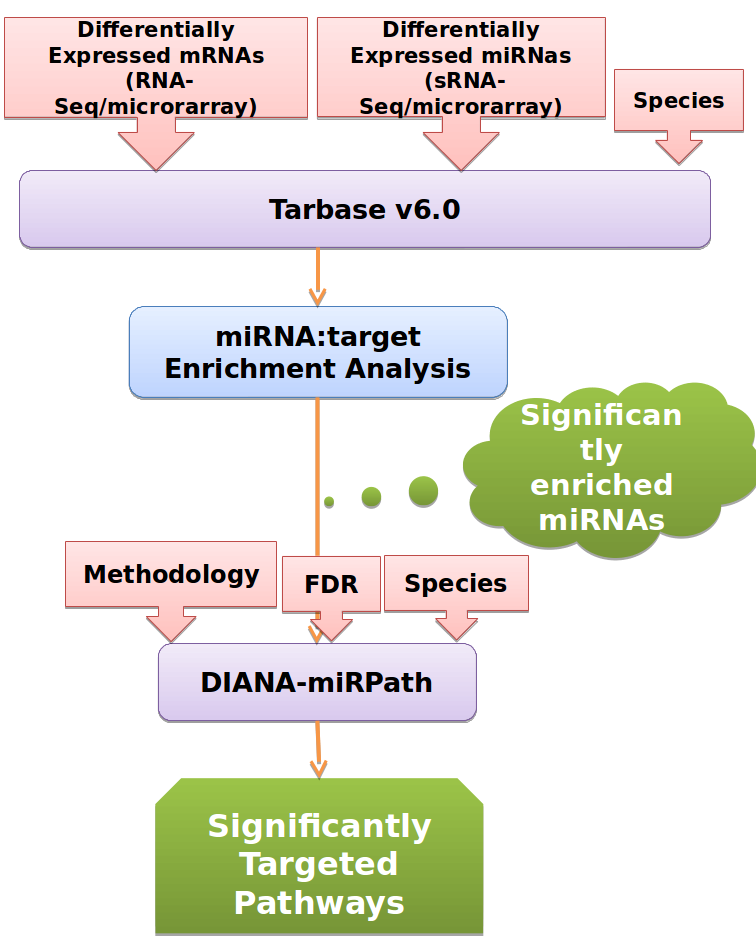
1.Example Workflow, performing enrichment analysis of validated miRNA:gene interactions followed by targeted Pathway analysis
The implemented workflow initially performs enrichment analysis in experimentally validated targets derived from DIANA-TarBase v6.0, identifying miRNAs significantly controlling the set(s) of differentially expressed genes. Subsequently a miRNA-targeted pathway analysis is implemented with DIANA-miRPath v2.
This Workflow makes use of DIANA-LAB services through the DIANA-web-server Plug in. In order for this workflow to work properly the plug-in has to be installed (Taverna v2.3 or v2.5).
Depending on the Taverna version you use, download one of the following files:
http://62.217.127.8/DianaTools/data/workflows/2.3/tarbase_v23.t2flow
or
http://62.217.127.8/DianaTools/data/workflows/2.5/tarbase_v25.t2flow
and open the workflow in the Taverna Workflow Management System.
The workflow can run automatically, , as soon as the necessary input values are provided. This is an active example of how DIANA-LAB services can be combined
in a single Workflow, and can be expanded utilizing other Taverna available Services.
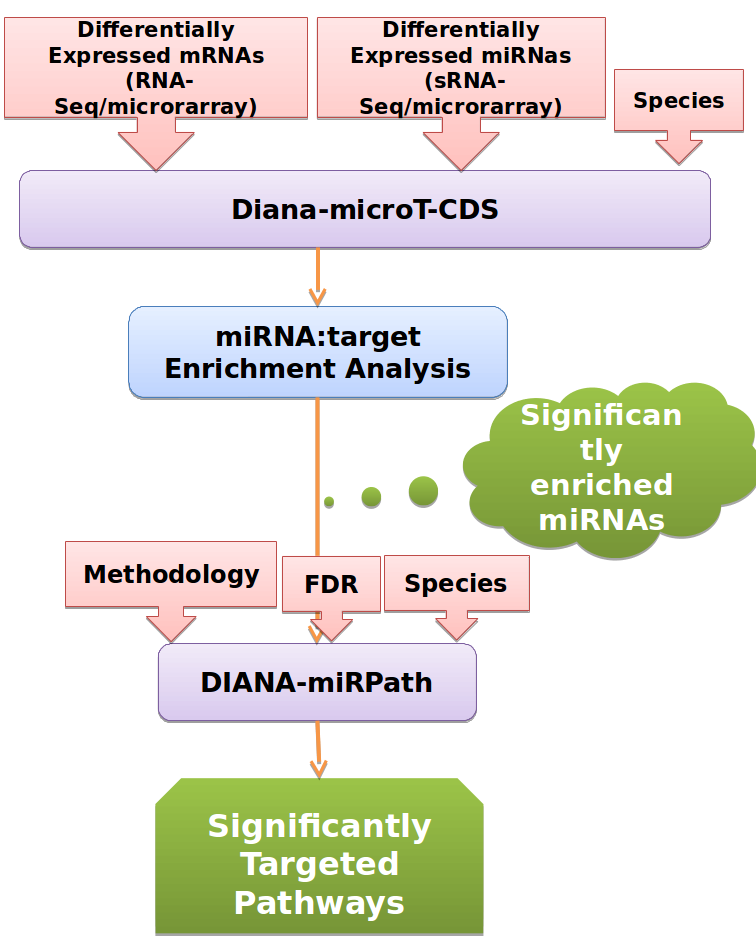
2. Example Workflow, performing enrichment analysis of predicted miRNA:gene interactions followed by targeted Pathway analysis
The implemented workflow initially performs enrichment analysis in in-silico predicted targets derived from DIANA-microT-CDS, identifying miRNAs significantly controlling the set(s) of differentially expressed genes. Subsequently a miRNA-targeted pathway analysis is implemented with DIANA-miRPath v2.
This Workflow makes use of DIANA-LAB services through the DIANA-web-server Plug in. In order for this workflow to work properly the plug-in has to be installed (Taverna v2.3 or v2.5).
Depending on the Taverna version you use, download one of the following files:
http://62.217.127.8/DianaTools/data/workflows/2.3/microt_v23.t2flow
or
http://62.217.127.8/DianaTools/data/workflows/2.5/microt_v25.t2flow
and open the workflow in the Taverna Workflow Management System. The workflow can run automatically, as soon as the necessary input values are provided.
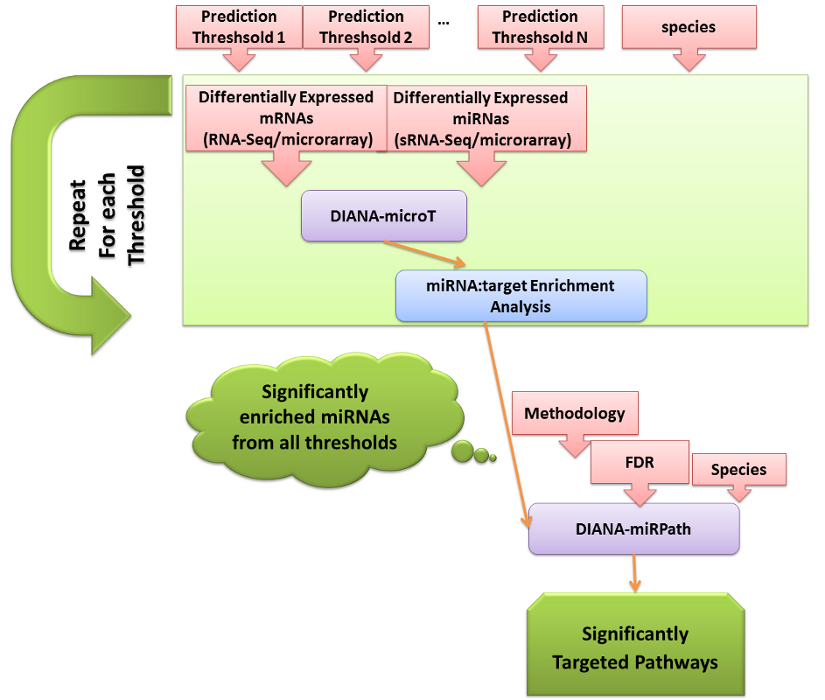
3. Example Workflow, integrating DIANA-LAB services. Optimized enrichment analysis of predicted miRNA:gene interactions followed by targeted Pathway analysis.
Interactions between user defined miRNA and gene sets are in silico identified using DIANA-microT-CDS. A subsequent miRNA target enrichment analysis identifies miRNAs controlling significantly the sets of differentially expressed genes. The pipeline is automatically repeated for different prediction thresholds (from sensitive to more stringent), in order to minimize the effect of the selected settings to the result. By utilizing meta-analysis statistics, the server combines the p-values from each repetition into a total p-value for each miRNA, signifying its effect on the selected genes for all utilized thresholds. In the last step of the pipeline, the identified miRNAs are subjected to a functional analysis with DIANA-miRPath v2, where pathways controlled by the combined action of these miRNAs are detected.
This Workflow makes use of DIANA-LAB services through the DIANA-web-server Plug in. In order for this workflow to work properly the plug-in has to be installed (Taverna v2.3 or v2.5).
Depending on the Taverna version you use, download one of the following files:
http://62.217.127.8/DianaTools/data/workflows/2.3/advanced_2_3.t2flow
or
http://62.217.127.8/DianaTools/data/workflows/2.5/advanced_2_5.t2flow
and open the workflow in the Taverna Workflow Management System. The workflow can run automatically, as soon as the necessary input values are provided.
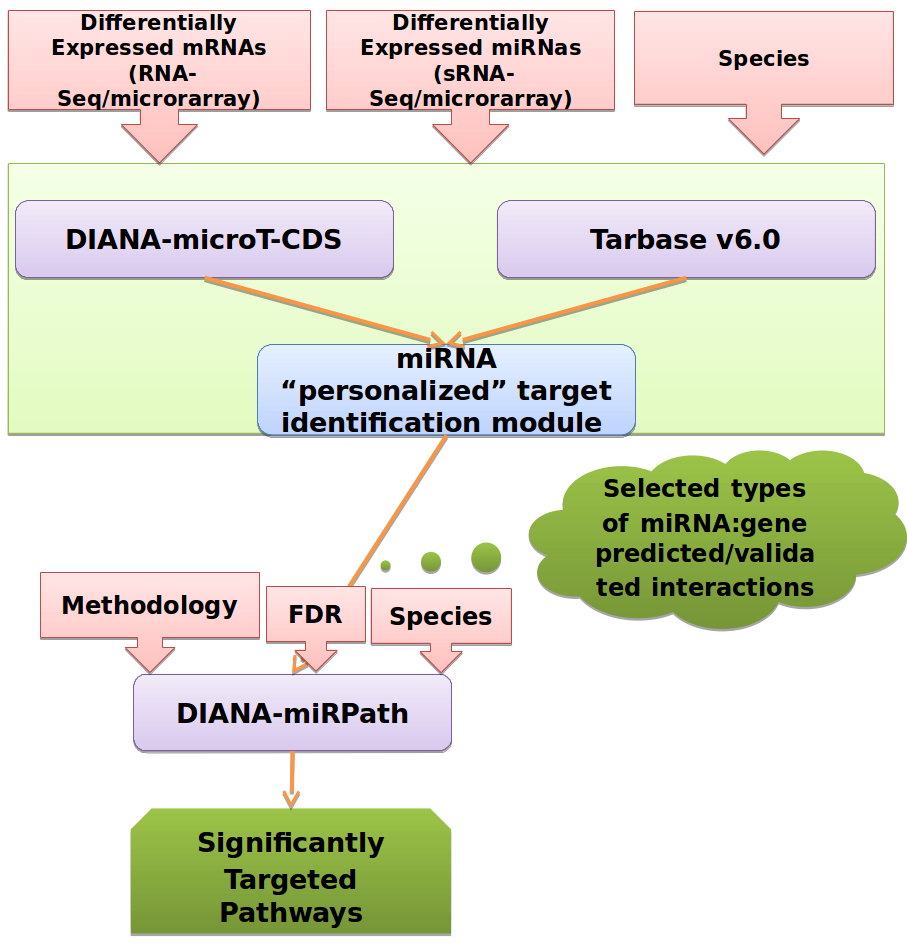
4. Example Workflow, “personalizing” the selection of miRNA-specific validated/predicted interactions, followed by miRNA Pathway analysis.
In this workflow, the algorithm "personalizes" the target identification module for each miRNA. It initially identifies the number of available interactions in DIANA-TarBase and DIANA-microT-CDS (validated vs predicted) and automatically selects to use validated targets only in the cases of well-annotated miRNAs. Computationally identified interactions are used otherwise. In the last step of the pipeline, the miRNAs with targets predicted form DIANA-microT-CDS or experimentally verified targets from TarBase v6 are subjected to a functional analysis with DIANA-miRPath v2, where pathways controlled by the combined action of these miRNAs are detected.
This Workflow makes use of DIANA-LAB services through the DIANA-web-server Plug in. . In order for this workflow to work properly the plug-in has to be installed (Taverna v2.3 or v2.5).
Depending on the Taverna version you use, the workflows can be downloaded from one of the following links:
http://62.217.127.8/DianaTools/data/workflows/2.3/tarbase_microt_v23.t2flow
or
http://62.217.127.8/DianaTools/data/workflows/2.5/tarbase_microt_v25.t2flow
The workflow can run automatically, as soon as the necessary input values are provided.
| |